Postprocessing¶
Postprocessing refers to evaluations and checks performed on a model to get an understanding of its properties and estimate its quality.
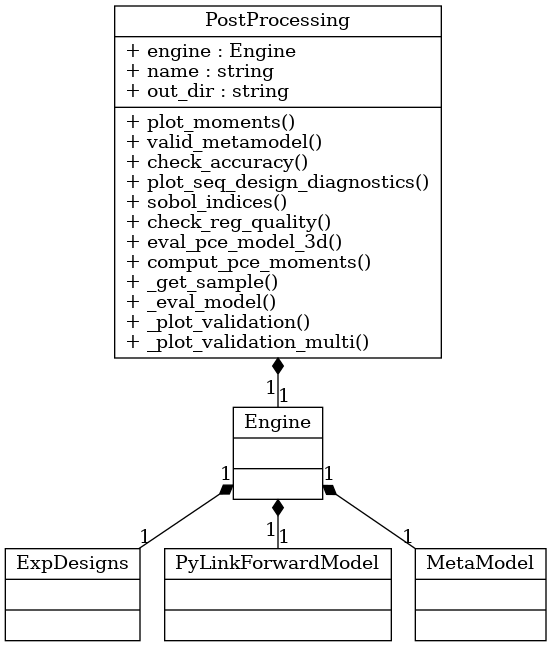
The BayesValidRox class bayesvalidrox.post_processing.post_processing.PostProcessing includes functions for evaluating and visualizing surrogate models.
plot_expdesign: Visualizing the set marginals and training pointsplot_moments: Visualizing the moments of the outputs of a surrogate modelvalidate_metamodel: Computing validation scores and visualizing the scores and the surrogate outputs; this function also callsplot_correl,plot_seq_design_diagnostics,plot_validation_outputsandplot_residual_hist
Postprocessing methods that are specific to a type of surrogate or training include
sobol_indices: Calculating the Sobol’ and Total Sobol’ indices (PCE, aPCE)
Example¶
We want to compare out trained surrogate from Active learning: iteratively expanding the training set against its original model using the class bayesvalidrox.post_processing.post_processing.PostProcessing.
>>> from bayesvalidrox import PostProcessing
The postprocessing object expects the full engine. This allows it to perform both sampling, running the model and running the surrogate model.
>>> post = PostProcessing(engine, out_format='png')
We start by visualizing the set marginals and the training points.
>>> post.plot_expdesign(show_samples = True)
If not specified otherwise, the plots are stored in the folder Outputs_PostProcessing_calib.

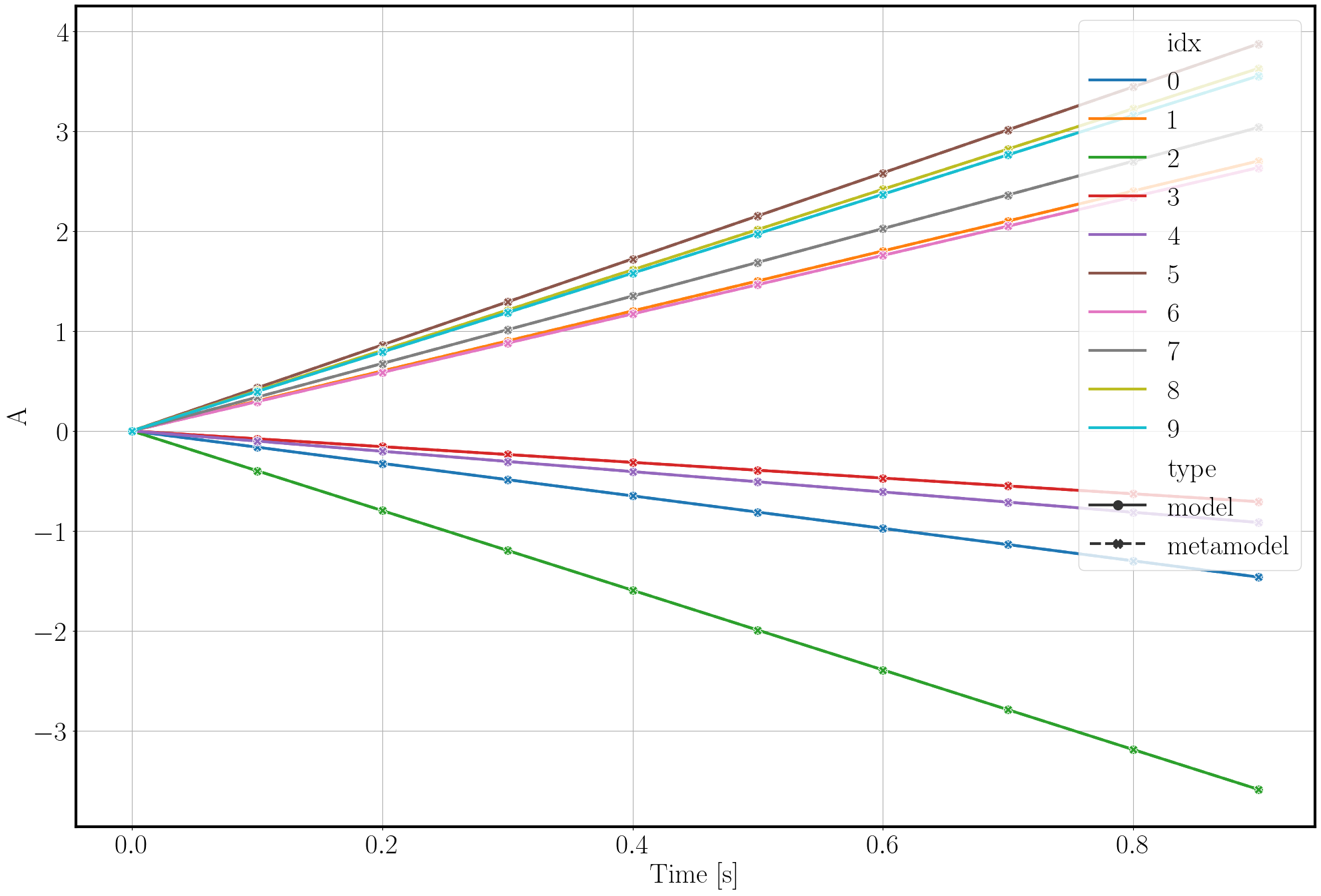
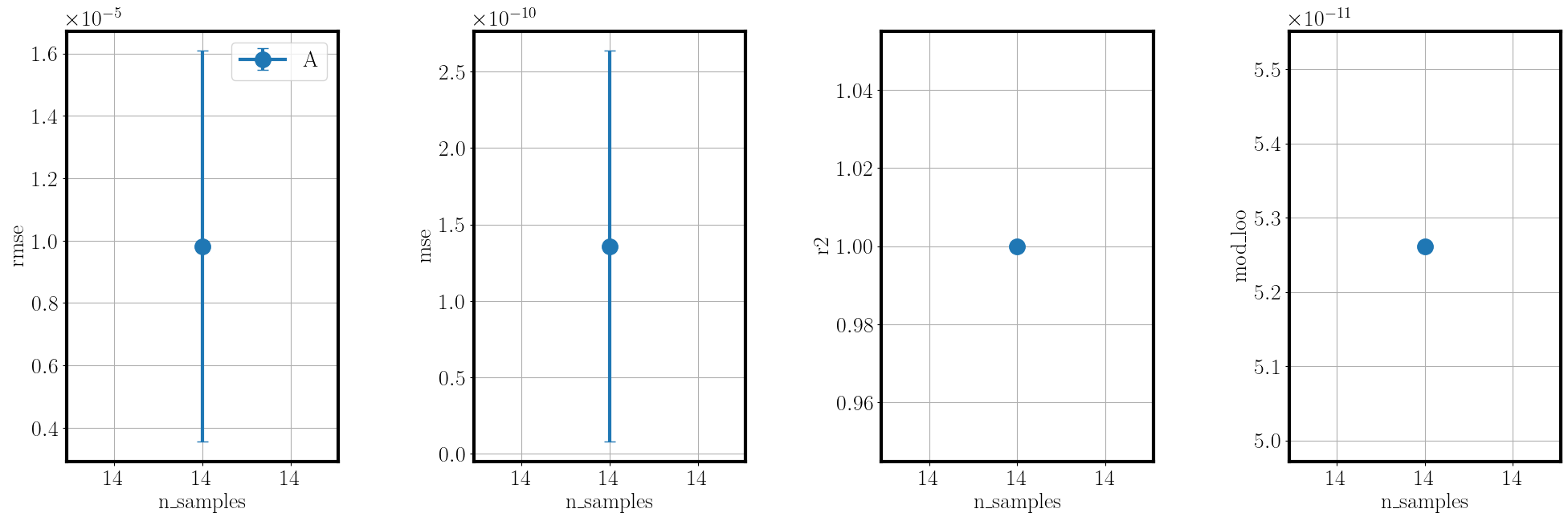
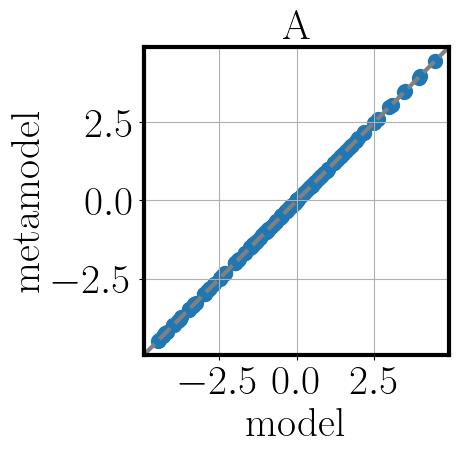
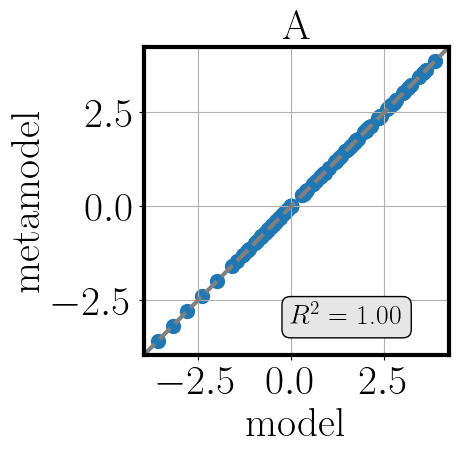
To gain an understanding of the approximation quality we use the function validate_metamodel.
Tihs function generates validation samples, then applies the applicable validation metrics and visualizes the results.
The validation metrics are selected based on the available data and include the RMSE, MSE and R2 values, mean and standard deviation errors of the moments against references, and Bayesian metrics such as BME, DKL and information entropy, if a reference observation is given.
>>> post.validate_metamodel(n_samples=10)
The visual comparison of model and metamodel results is saved in the folder Outputs_PostProcessing_calib.